Exploring intein structures in SPDBV
Background information:
Most inteins are composed of two domains: one
is responsible for protein splicing, and the other has
endonuclease activity. A few inteins have lost the
endonuclease domain completely and retain only the
self-splicing domain and activity. The latter inteins are called mini-inteins.
The structures of several inteins are crystallized.
- Saccharomyces cerevisiae intein (PMID: 9160747, 1VDE),
- the same Saccharomyces cerevisiae intein bound to its recognition sequence
(PMID: 12219083, 1LWS),
- the Mycobacterium xenopi mini intein (PMID: 9437427, 1AM2)
- Open 1VDE in SPDBV. This structure has two chains. Save chain A to a separate
file.
- Reopen chain A in SPDBV. Open Mycobacterium mini intein. Align two structures
using Magic Fit. How good is the alignment? (your answer should be "Bad").
Depict the structures as ribbons and color them according to the secondary
structure. Rotate two structures until you can see the similarities between
mini intein and one of the domains of large intein. Move them on top of each
other. Do "Fit -> Improved Fit...". Does it work now?
Can you find which part of Saccharomyces cerevisiae intein corresponds to
the endonuclease domain by comparison of the two structures? [Manually inspect
the two structures to find the similarities]. Color the putative self-splicing
and endonuclease domains of 1VDE in two different colors. Select N and C terminals
(First a.a. and the last a.a.) in both structures. How close are they (in
angstroms)? [Click on the button with "1.5A" label, and select first and second
a.a. to obtain the distance]. Save your project.
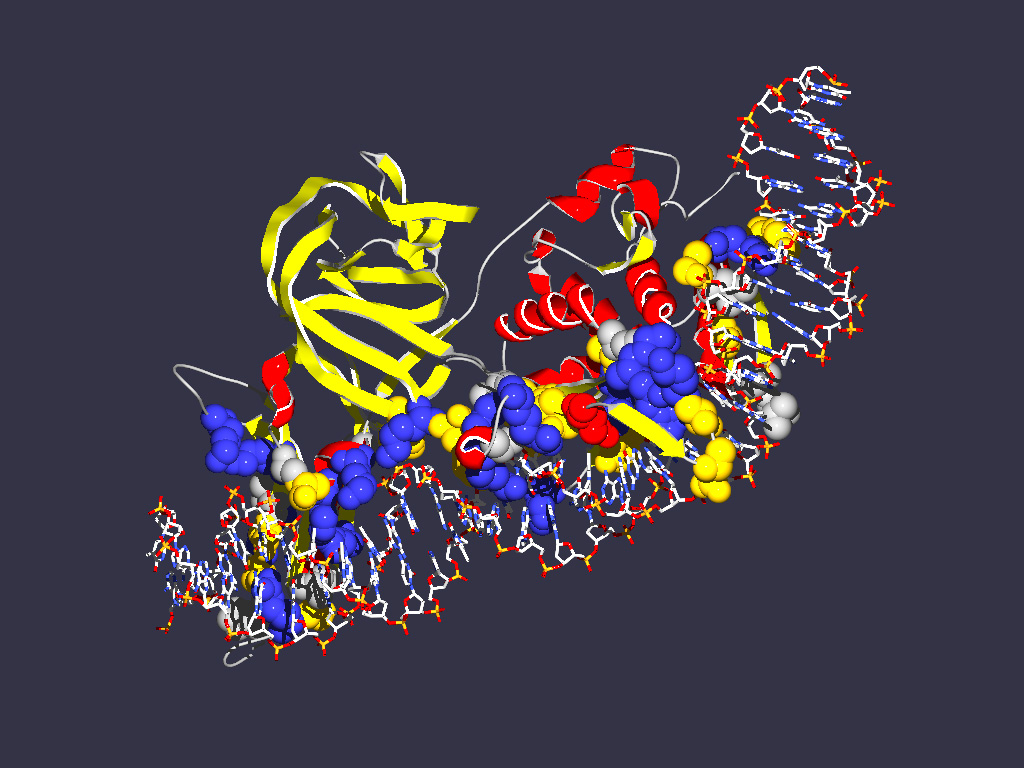
- Open Saccharomyces cerevisiae intein that is bound to its recognition
DNA sequence. Display intein (chain A) as ribbons with secondary structure
color scheme (select color target with little black triangle in Control Panel).
Color DNA as CPK, and compute hydrogen bonds (Tools - > compute H-bonds).
Does the finding of DNA interaction domain agree with your previous assignment
of the self-splicing domain? (see the saved structure from the previous exercise)
- Try to find a way to display the interactions between the aminoacid side
chains and the DNA helix. One way to do it is to select two DNA chains and
select Neighbors of selected amino acids. Choose "select groups that are within"
option. Click on the "Cloud" icon in the header of the control panel to display
the aminoacids as balls. Also click in the header for showing sidechain. Manually
turn off the cloud checkmarks for the DNA. One way to look at individual interactions
is to turn the molecule so that one looks down the DNA helix, and select the
"Display -> Slab" option. If you press shift while mouse cursor up and down
the visible slice moves through the molecule along the axes perpendicular
to the screen. If you press the shift key and move the mouse left and right
you increase or decrease the size of the slab.
- The Lys 340 and Glu 366 are residues that are important for interaction
with DNA. Select those residues (in Control panel choose label column to depict
the label). Do they interact with major or minor groove of DNA? Which base
pairs interact with these amino acids?