Aligning F-ATPase alpha and
beta subunits
Start SPdbV
Open 1bmf.pdb
Color Chain
Change color chainD to
grey/blue (rightclick in control panel on D in first column to select chain D,
right click on COL, select color)
Scrol down the control
panel and select all ATP analogs (press ctrl key and right click to select)
right click on COL in
heading and select red color
Read the pdb file to get
info on which chain is which
select chain F (including
nuc) and save selected residues as betaTP.
select chain A (including
nuc) and save selected residues as alphaE.
After playing with the
F1-ATPase, close this file and open betaTP and alphaE.
Display layer info
select and display only the
nucleotides
There are
different ways to align 3-D structures. One way is to select 3 corresponding
points in each of the two structures. To do so you can use the substrate
molecule.
Using the mov check off in
the Layer Info, reorient the two ANPs so that they are in a similar orientation
(but not overlapping).
Click on the align bottom
with the 3 green and 3 red dots. Notice the red instructions that appear in the
header next to the pdb-page icon. Follow these instruction using three
corresponding atoms.
SHIFT DISPLAY CA chain
(Shift makes the commands act on both layers)
Using the mov checks in the
Layer info, move the two chains next to each other.
What do you think about the
result?
Another way to align
structures is to use the magic fit in the tools command. Do this and run
improve fit (notice the red info in the header)
Click on alpha in Layer
info to make the alpha subunit the active layer
Color CPK
Make the beta subunit the
active layer
COLOR rms . The further the
atoms in the beta subunit are away from the alpha subunit, the longer
wavelengths it is the colored.
DISPLAY Show alignment
window - gives you the aligned sequences.
How does this alignment
compare to the ones calculated using Clustalw?
If you
have time, repeat the exercise for the three beta subunits to observe
the structural changes the beta subunit is undergoing in the catalytic cycle.
If you have more time to
spare and you are up for a challenge, take a look at the nucleosome.
Right click here and save as pdb file.
Open it from within spdbv. You
might want to do some of the future exercises with the nucleosome in addition
to the ATPases – thus save the pdb file, where you can find it again.
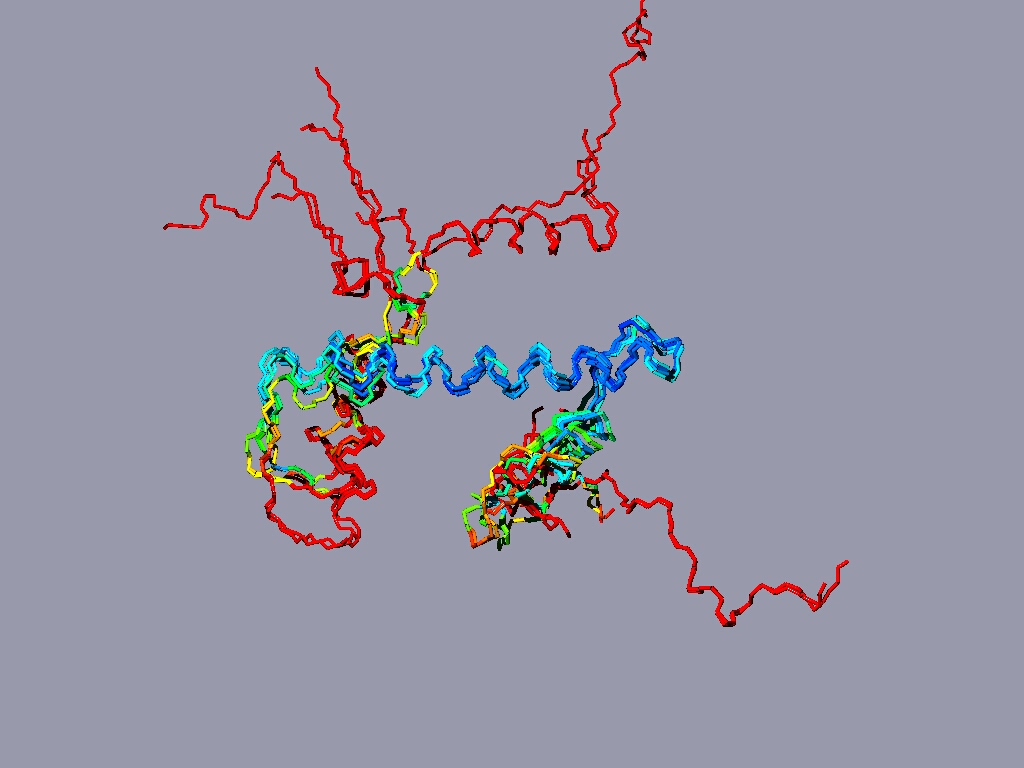
Align all the histones form the nucleosome to one reference histone and color in rmv:
The result might look something like this:
The picture shows a structure alignment of the 8 histones (2 each) that are part of the nucleosome. All the histones were colored regarding the match to H2A, except H2A, which was colored according to its match to H3. Coloring option RMS – shorter wavelengths – better match
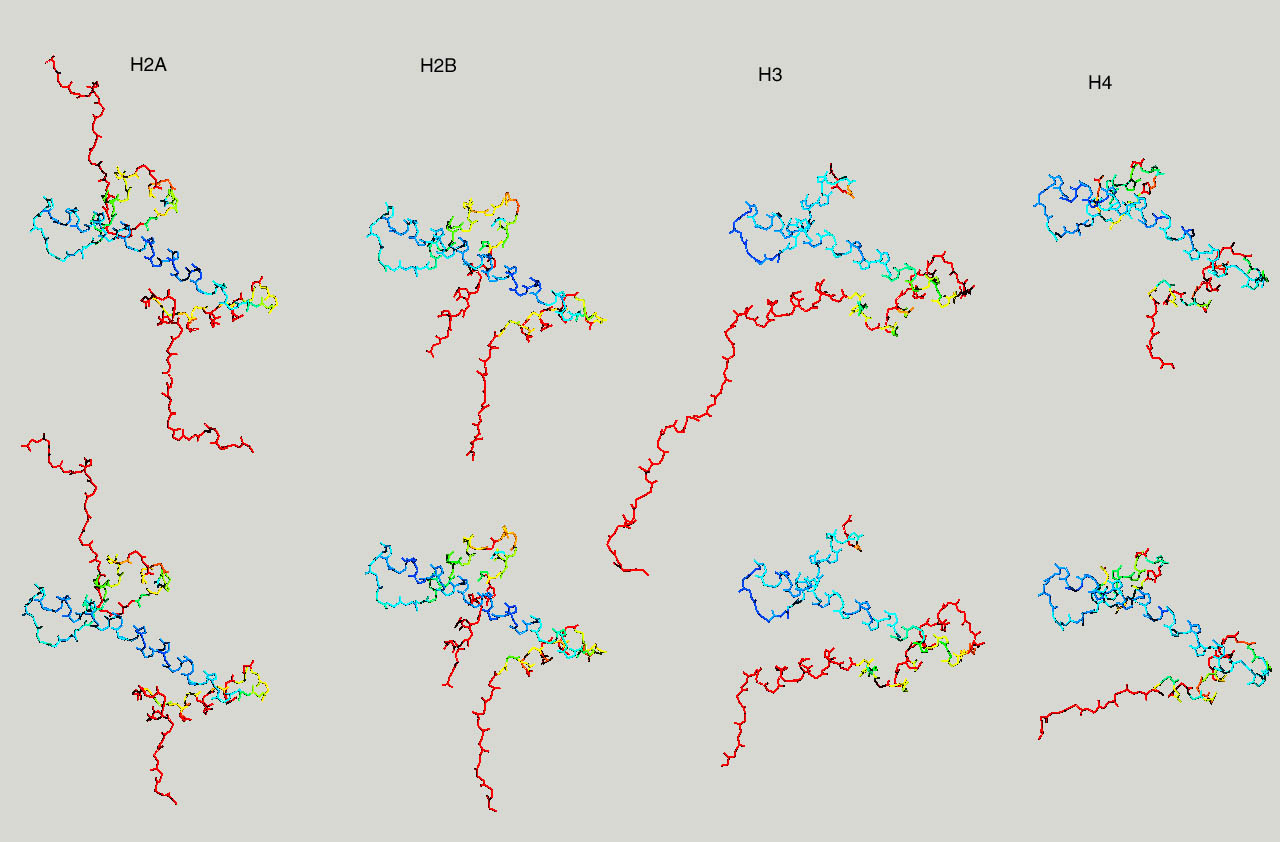
Below same as last figure, but histones are depicted side by side
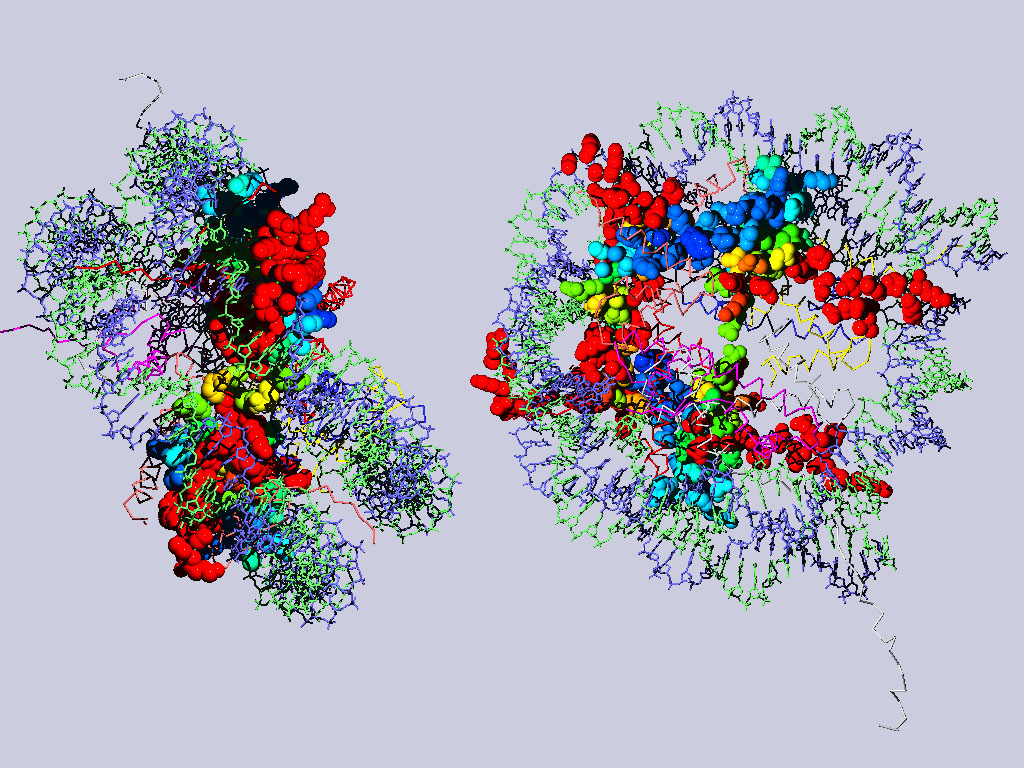
Below are two views of the complete nucleosome. Histones H2A are depicted as spacefilling balls and RMS colored regarding their match to H3. The rest of the molecule is colored according to chain.