Optional exercises:
Glutathione synthetase and D-Alanine D-Alanine Ligase were long ago (1990) recognized by Jim Knox (MCB faculty) to be so similar in structure that there could be no doubt about their common evolutionary origin. Later these and other enzymes were discovered to have a novel ATP binging site (the GRASP domain). These domains were identified using profile alignments (will be covered later in this course). A description of the GRASP domain family is at https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3249071/.
Save the following files to your computer and load them into chimera.
2DLN.pdb (D-Alanine D-Alanine Ligase - D-ala is an important part of the bacterial cell wall, more here
1GSA.pdb (glutathione synthetase from E. coli, glutathione is the biological equivalent of mercaptoethanol),
cpsBfrag.pdb,
load cpsFfrag.pdb
(Carbamoyl
phosphate synthetase is an enzyme consisting of several domains.
These are the front and the back of (1BXR)).
Based on your first impression, are these structures similar? homologous?
| Your answer ---> |
Can you use Tools > Structure comparison > Matchmaker to align these structures?
| Your answer ---> |
See here for an illustration of the structures in similar orientation.
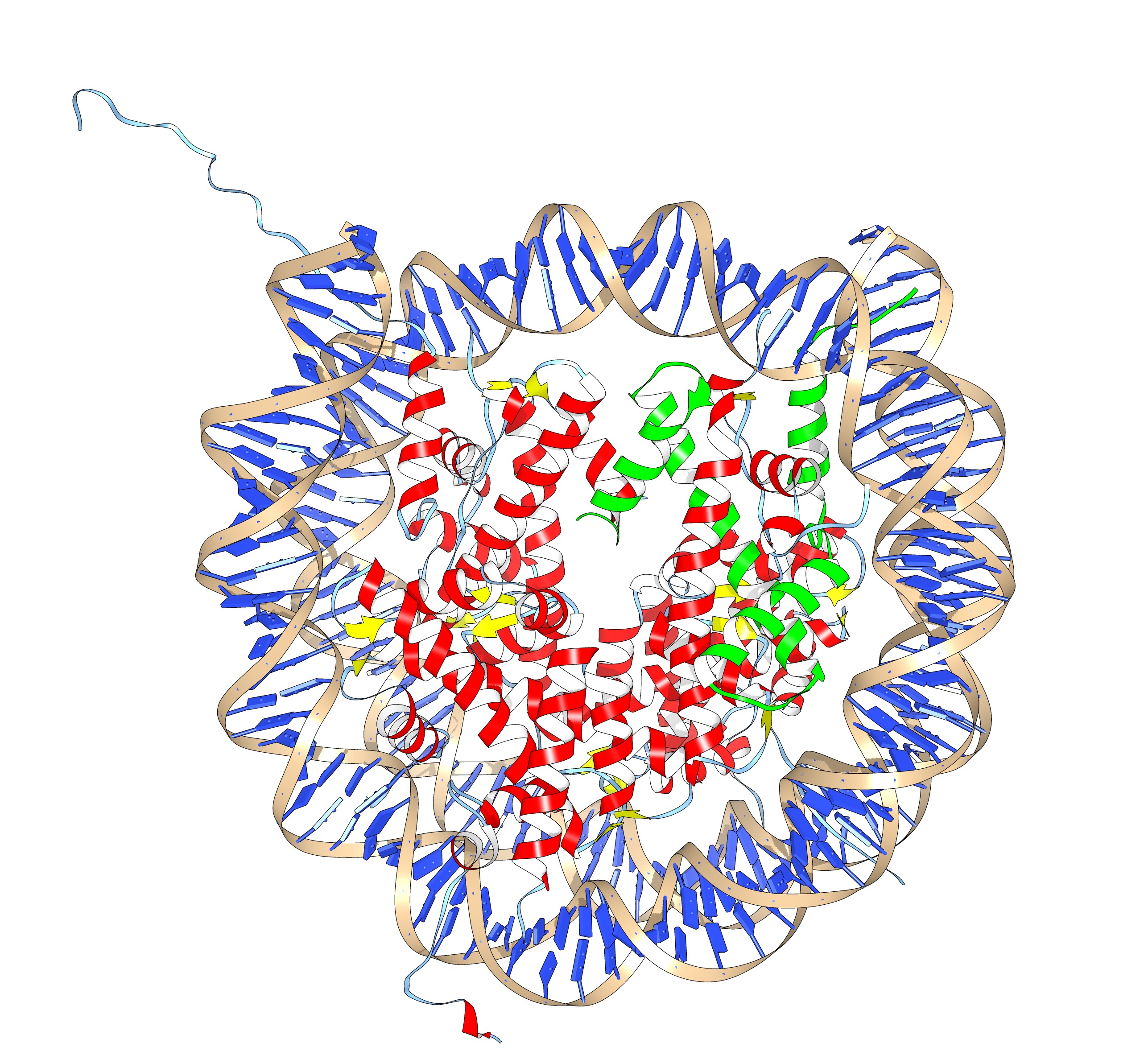
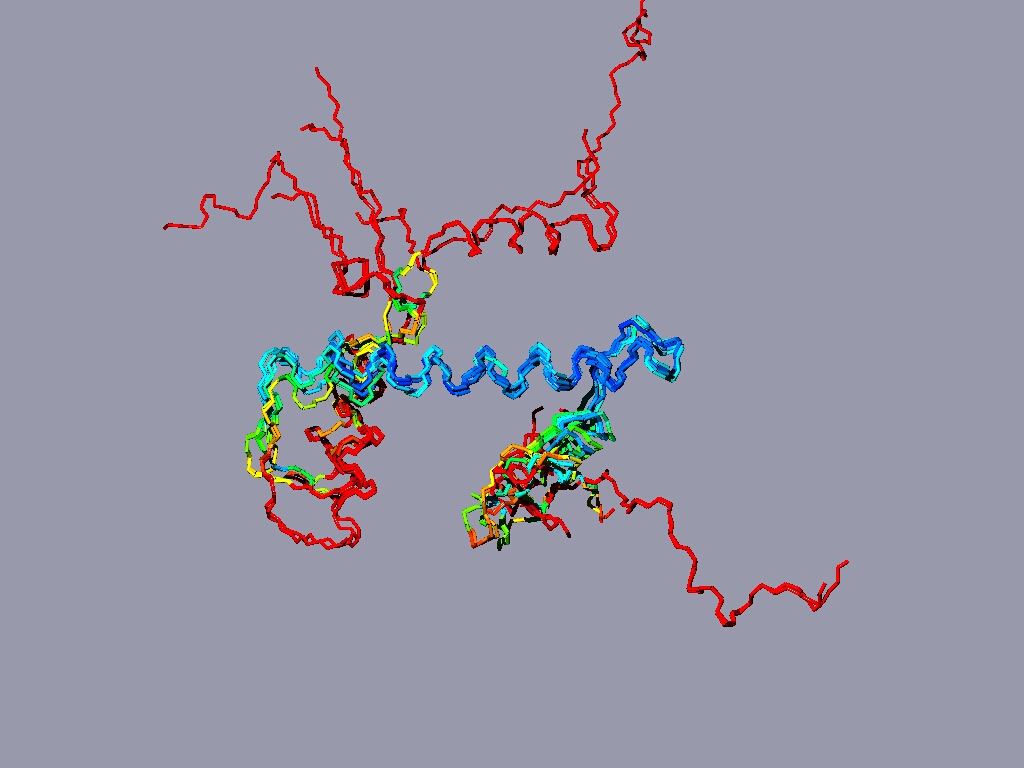
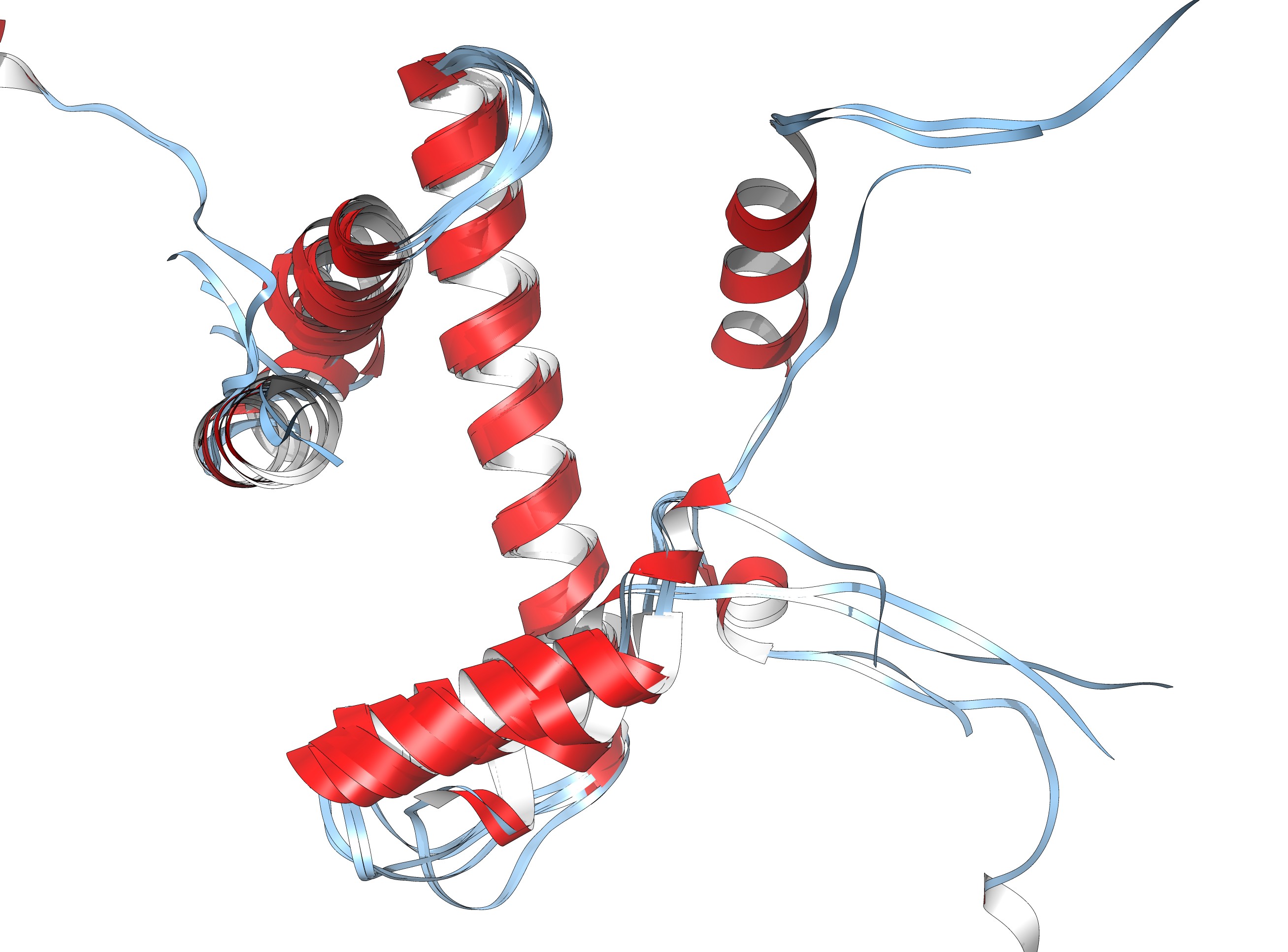
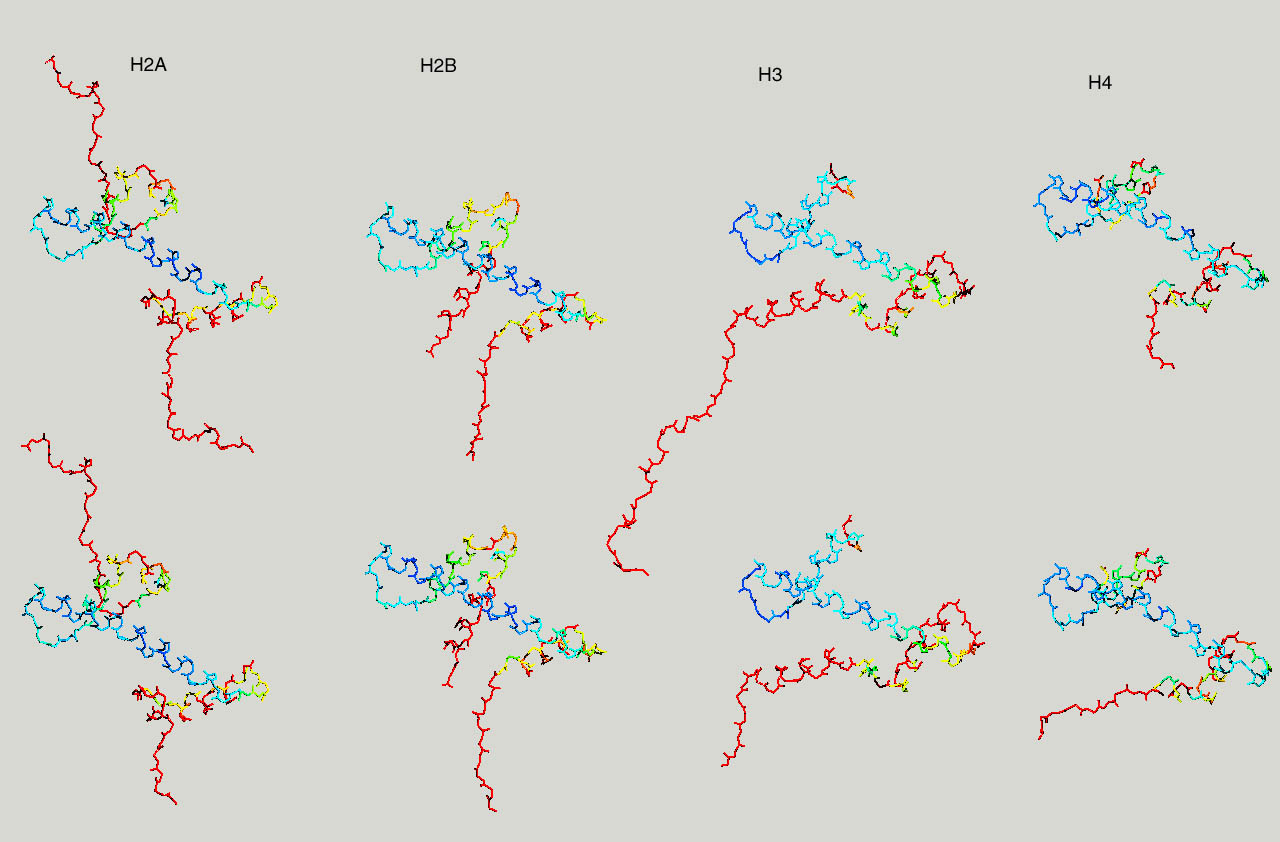
If you have more time to spare and you are up for a challenge, take a look at the nucleosome (1AOI.pdb). Open it from within chimera. Save each of the 8 histones as a separate pdb file. Close the nucleosome file, open the 8 histone files and align them to one reference histone.
The nucleosome, one of the 8 histones is colored green. (2 copies of each paralog)

The eight histones aligned to one another.
Same as ribbon display
Below same as last figure, but histones are depicted side by side :
Proteasomes are the recycling cans of the cell. Ubiquitin tagged protein are taken-up into the proteasome and broken down into aa. The proteasome (20S) core consists out of homologous alpha and beta subunits, arranged in 4 rings. The two central ones are beta subunits (2x7), the two rings at the ends are made from alpha subunits. Humans (and other eukaryotes) have 7 different genes encoding seven different alpha subunits, and 7 different genes encoding beta subunits, and each is present in duplicate in the 20S proteasome core. Altogether 28 subunits.
We are interested in creating a structure based sequence alignment of the different alpha and beta subunits. To do so, pick the bottom or top half of the proteasome (5LF7.pdb, or a similar high resolution structure from another organism), and save all the different subunits as separate pdb files (if you use 5LF7.pdb, the pdbs for the individual SU are here). First align all the alpha beta subunits separately, and then try to generate an alignment of the alpha with the beta subunits. Use matchmaker to align the structures, then use Tools > structure comarison > Match-Align to create a multiple sequence alignment.
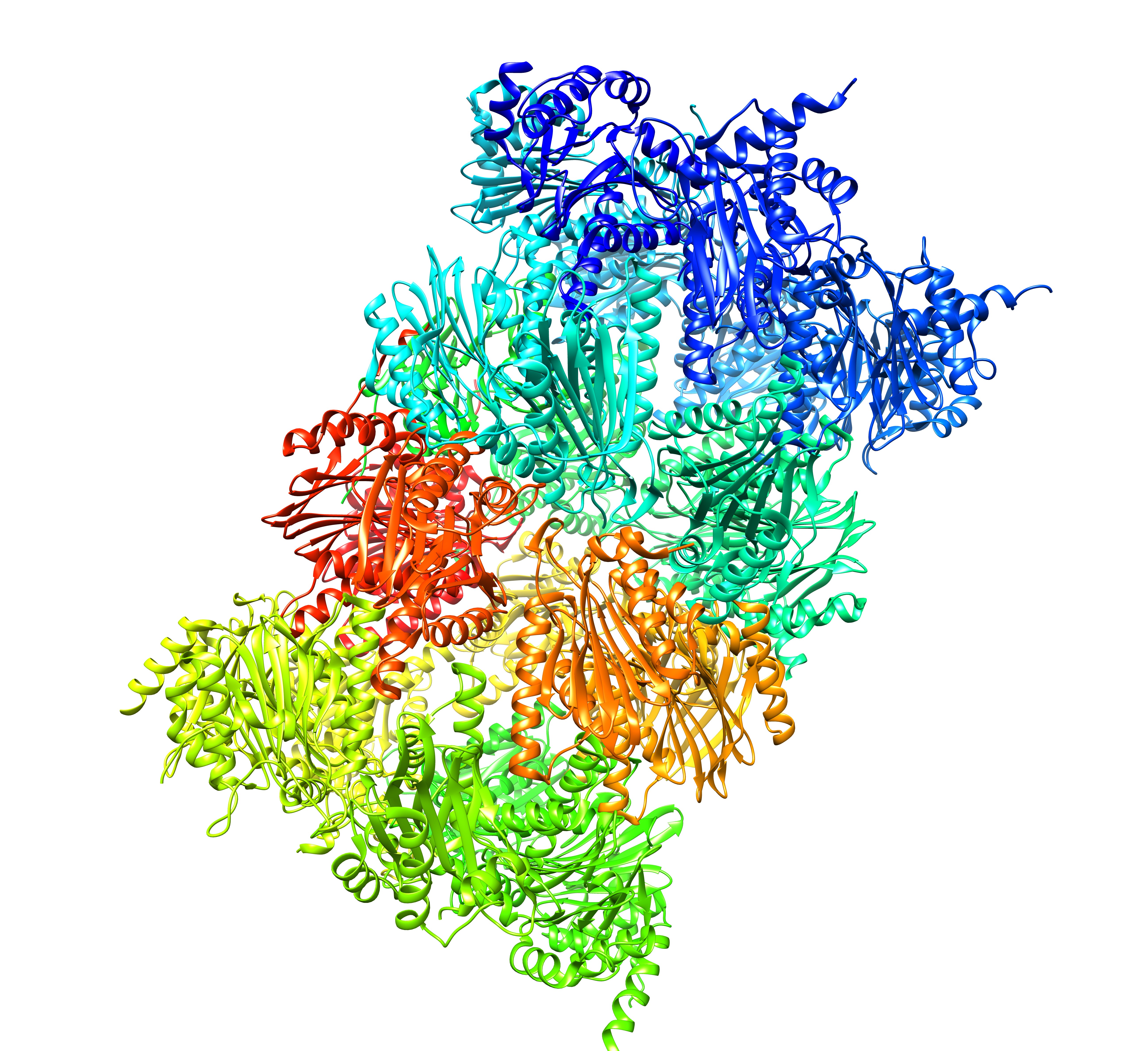
The human 20S proteasome core (28 subunits, 14 paralogs)
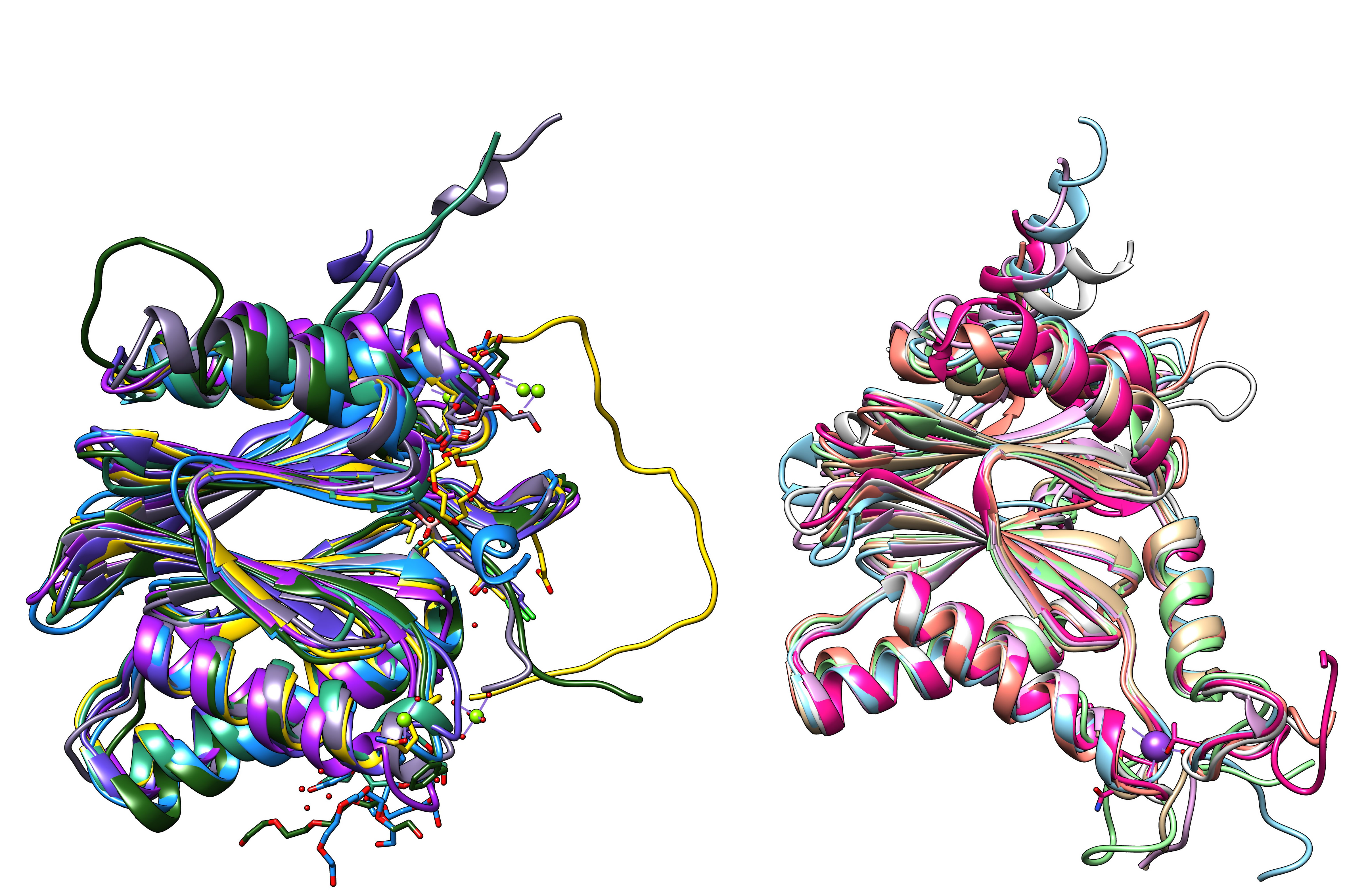
Proteasome subunits (7 beta and 7 alpha subunits) from human hela cells (5LF7.pdb)

All 14 subunits of the human proteasome core superimposed.
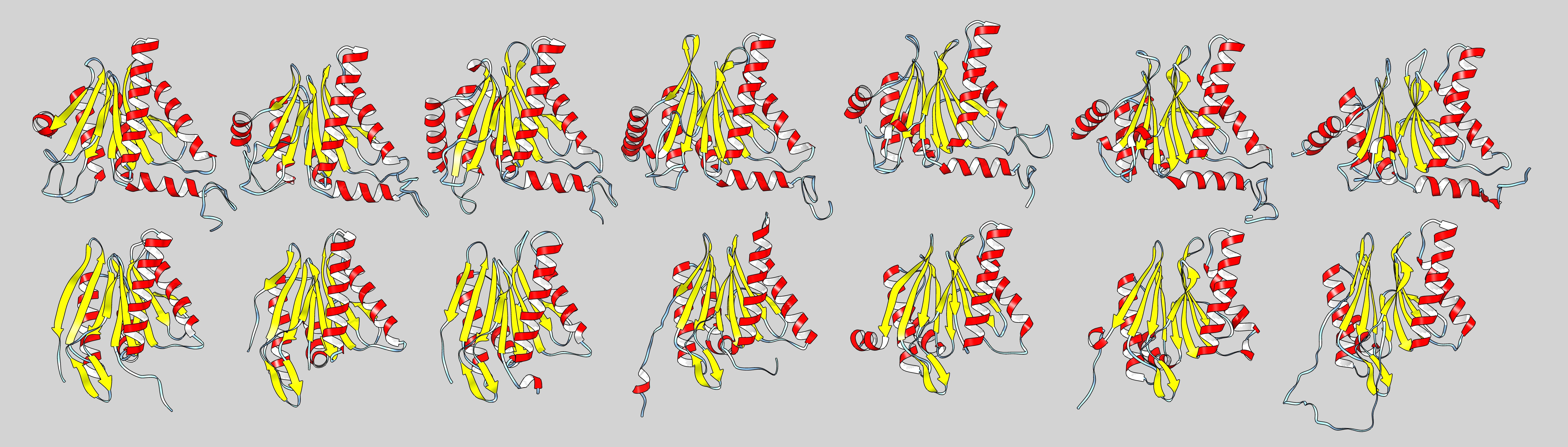
All 14 subunits of the human proteasome next to one another